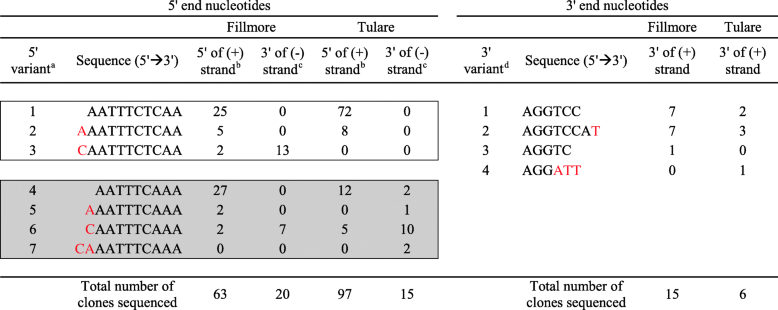
- aThe extreme 5′ end sequences of seven nt variants were identified by sequencing cloned DNA corresponding to the 5′ end of the T36-CA genome. In comparison with the T36 consensus genomic 5′ end nt sequences (black colored), the nt variants of T36-CA were found to contain extra/variant nt (s) (red colored nts). Group 1 nt variants (variants 1 to 3) (unshaded box) have consensus 5′ nt sequence “AATTTCTCAA” identical to that of T36 isolate FS674 (GenBank accession no. KC517485) and FS703 (GenBank accession no. KC517487). Group 2 nt variants (variants 4 to 7) (shaded box) have consensus 5′ nt sequence “AATTTCAAA” similar to that (“AATTTCACAA”) of the T36 sequences (GenBank accession nos. EU937521 and AY170468), except that it does not contain a “C” at position 8. The number of clones for each nt variant sequenced are as indicated
- bThe 5′ end nts of the T36-CA genome was determined by sequencing the (+)-DNA generated by 5′ RLM-RACE using the (+)-strand of dsRNA as a genetic template for reverse transcription (RT) and PCR amplification
- cThe 5′ end nts of the T36-CA genome was determined by 3′ RACE using the (−)-strand of dsRNA as a genetic template for reverse transcription (RT) and PCR amplification
- dThe extreme 3′ end sequences of 4 nt variants were identified by sequencing cloned (+)-DNA (generated by 3′ RACE) corresponding to the 3′ end of the T36-CA genome. The T36 consensus genomic 3′ end nts (black colored) and the T36-CA nt variants (red colored) are as shown. The number of clones for each nt variant sequenced are as indicated

